COVID-19
COVID-19 Pandemic in South America
View the Project on GitHub bgonzalezbustamante/COVID-19-South-America
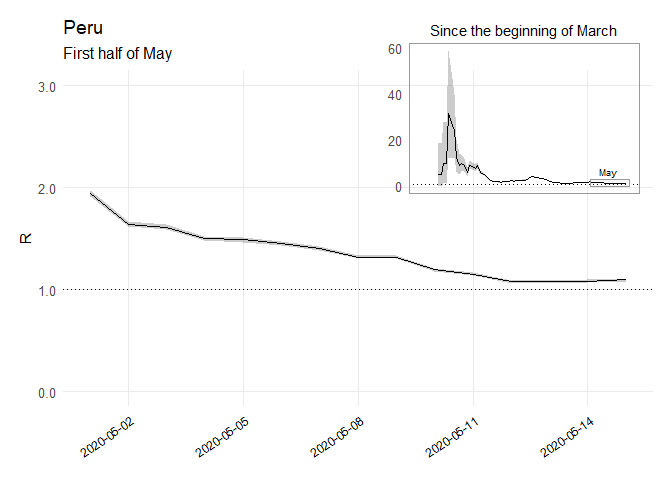
Parametric Re Estimations Peru Code
## Dataframe
data_per <- select(south_am, dates, PER)
names(data_per)[2] = "I"
## Parametric Re
per_param_si <- estimate_R(data_per, method="parametric_si",
config = make_config(list(
mean_si = 7.5, std_si = 3.4)))
## Plot 1
per_Re_full <-
plot(per_param_si, "R") + theme_minimal(base_size = 12) +
theme(legend.position = "none") +
theme(panel.grid.minor = element_blank(),
panel.grid.major = element_blank(),
panel.border = element_rect(color = "grey60", size = 0.5, fill = NA),
plot.title = element_text(size = 11, hjust = 0.5, vjust = 2.5,
margin = margin(), color = "black")) +
theme(axis.text.x = element_blank()) +
labs(x = NULL, y = NULL, title = "Since the beginning of March",
subtitle = NULL, caption = NULL) +
theme(plot.margin = unit(c(0.5,0.5,0.5,0.5), "cm")) +
scale_y_continuous(labels = scales::comma) +
theme(axis.title.y.left = element_text(size = 11)) +
annotate("rect", xmin = as.Date("2020-05-01"), xmax = as.Date("2020-05-16"),
ymin = 0, ymax = 3, alpha = .05, colour = "grey60") +
annotate(geom = "text", x = as.Date("2020-05-8"), y = 3 * 2.1,
label = "May", size = 2.5)
## Plot 2
per_Re_may <-
plot(per_param_si, "R") + theme_minimal(base_size = 12) +
theme(legend.position = "none") +
theme(panel.grid.minor = element_blank()) +
theme(axis.text.x = element_text(angle = 35, hjust = 1, color = "black",
size = 9)) +
labs(x = NULL, y = "R", title = "Peru",
subtitle = "First half of May", caption = NULL) +
theme(plot.margin = unit(c(0.5,0.5,0.5,0.5), "cm")) +
scale_y_continuous(labels = scales::comma, limits = c(0, 3)) +
scale_x_date(date_breaks = "3 days", date_minor_breaks = "3 days",
date_labels = "%Y-%m-%d",
limits = c(as.Date("2020-05-01"), as.Date("2020-05-15"))) +
theme(axis.title.y.left = element_text(size = 11))
## Integration
ggdraw() +
draw_plot(per_Re_may) +
draw_plot(per_Re_full, x = 0.55, y = 0.55, width = .43, height = .43)