COVID-19
COVID-19 Pandemic in South America
View the Project on GitHub bgonzalezbustamante/COVID-19-South-America
Descriptive Analysis Code
Precoding
## Packages
library(EpiEstim)
library(incidence)
library(ggplot2)
library(scales)
library(shadowtext)
library(dplyr)
library(epitrix)
library(distcrete)
library(projections)
library(earlyR)
library(broom)
library(lubridate) ## PS-Note 2020-08-30: It had already been loaded.
library(grid)
library(cowplot)
library(tidyverse) ## PS-Note 2020-08-30: It had already been loaded.
library(lubridate) ## PS-Note 2020-08-30: It had already been loaded.
## Incidence
covid19 <- confirmed_covid19
names(covid19)[1] = "dates"
## Correction URY 12 April (2.87%)
covid19[47, 9] <- 0
## Deaths
names(deaths_covid19)[1] = "dates"
## Recovered
names(recovered_covid19)[1] = "dates"
## ICU Beds
icu_arg <- 8444
icu_bol <- 220
icu_bra <- 55101
icu_chl <- 2724
icu_col <- 5354
icu_ecu <- 1183
icu_pry <- 734
icu_per <- 721
icu_ury <- 700
## Country Names
arg_name <- "Argentina"
bol_name <- "Bolivia"
bra_name <- "Brazil"
chl_name <- "Chile"
col_name <- "Colombia"
pry_name <- "Paraguay"
per_name <- "Peru"
ury_name <- "Uruguay"
Descriptive Analysis
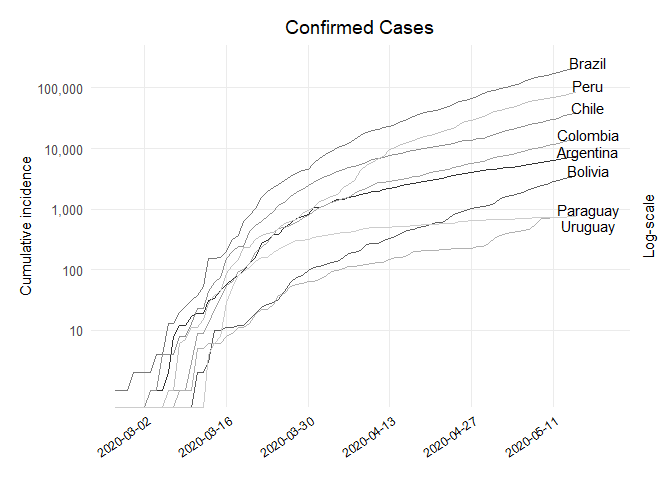
## Dataframe Confirmed
df_confirmed <- data.frame(dates = c(rep(as.Date(covid19$dates), 8)),
value = c(cumulate(covid19$ARG),
cumulate(covid19$BOL),
cumulate(covid19$BRA),
cumulate(covid19$CHL),
cumulate(covid19$COL),
cumulate(covid19$PRY),
cumulate(covid19$PER),
cumulate(covid19$URY)),
country = c(rep(arg_name, length(covid19$dates)),
rep(bol_name, length(covid19$dates)),
rep(bra_name, length(covid19$dates)),
rep(chl_name, length(covid19$dates)),
rep(col_name, length(covid19$dates)),
rep(pry_name, length(covid19$dates)),
rep(per_name, length(covid19$dates)),
rep(ury_name, length(covid19$dates))))
## Plot Confirmed
ggplot(df_confirmed, aes(x = dates, y = value, colour = country)) +
geom_line() + scale_colour_grey() + theme_minimal(base_size = 12) +
theme(legend.position = "none") + theme(panel.grid.minor = element_blank()) +
theme(axis.text.x = element_text(angle = 35, hjust = 1, color = "black", size = 9)) +
labs(x = NULL, y = "Cumulative incidence", title = "Confirmed Cases", subtitle = NULL,
colour = NULL) + theme(plot.margin = unit(c(0.5,0.5,0.5,0.5), "cm")) +
scale_x_date(date_breaks = "2 weeks", date_minor_breaks = "2 weeks",
date_labels = "%Y-%m-%d", limits = c(as.Date("2020-02-26"),
as.Date("2020-05-20"))) +
scale_y_log10(breaks = 10**(1:10), labels = comma(10**(1:10)),
sec.axis = sec_axis(~ ., labels = NULL, name = "Log-scale")) +
theme(axis.title.y.right = element_text(angle = 90, size = 11),
axis.title.y.left = element_text(size = 11),
plot.caption = element_text(size = 9),
plot.title = element_text(hjust = 0.5)) +
annotate("text", y = df_confirmed$value[which(df_confirmed$date == "2020-05-15"
& df_confirmed$country == "Argentina")]
* 1.2, x = as.Date("2020-05-17"), label = "Argentina") +
annotate("text", y = df_confirmed$value[which(df_confirmed$date == "2020-05-15"
& df_confirmed$country == "Bolivia")]
* 1.2, x = as.Date("2020-05-17"), label = "Bolivia") +
annotate("text", y = df_confirmed$value[which(df_confirmed$date == "2020-05-15"
& df_confirmed$country == "Brazil")]
* 1.2, x = as.Date("2020-05-17"), label = "Brazil") +
annotate("text", y = df_confirmed$value[which(df_confirmed$date == "2020-05-15"
& df_confirmed$country == "Chile")]
* 1.2, x = as.Date("2020-05-17"), label = "Chile") +
annotate("text", y = df_confirmed$value[which(df_confirmed$date == "2020-05-15"
& df_confirmed$country == "Colombia")]
* 1.2, x = as.Date("2020-05-17"), label = "Colombia") +
annotate("text", y = df_confirmed$value[which(df_confirmed$date == "2020-05-15"
& df_confirmed$country == "Paraguay")]
* 1.3, x = as.Date("2020-05-17"), label = "Paraguay") +
annotate("text", y = df_confirmed$value[which(df_confirmed$date == "2020-05-15"
& df_confirmed$country == "Peru")]
* 1.3, x = as.Date("2020-05-17"), label = "Peru") +
annotate("text", y = df_confirmed$value[which(df_confirmed$date == "2020-05-15"
& df_confirmed$country == "Uruguay")]
* 0.7, x = as.Date("2020-05-17"), label = "Uruguay")
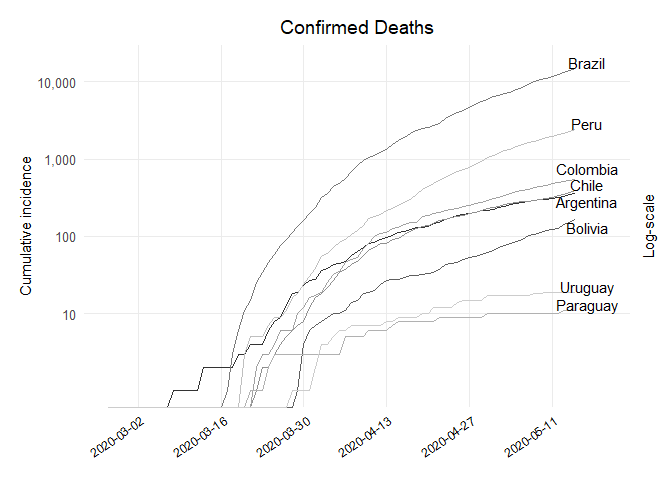
## Dataframe Deaths
df_deaths <- data.frame(dates = c(rep(as.Date(deaths_covid19$dates), 8)),
value = c(cumulate(deaths_covid19$ARG),
cumulate(deaths_covid19$BOL),
cumulate(deaths_covid19$BRA),
cumulate(deaths_covid19$CHL),
cumulate(deaths_covid19$COL),
cumulate(deaths_covid19$PRY),
cumulate(deaths_covid19$PER),
cumulate(deaths_covid19$URY)),
country = c(rep(arg_name, length(deaths_covid19$dates)),
rep(bol_name, length(deaths_covid19$dates)),
rep(bra_name, length(deaths_covid19$dates)),
rep(chl_name, length(deaths_covid19$dates)),
rep(col_name, length(deaths_covid19$dates)),
rep(pry_name, length(deaths_covid19$dates)),
rep(per_name, length(deaths_covid19$dates)),
rep(ury_name, length(deaths_covid19$dates))))
## Plot Deaths
ggplot(df_deaths, aes(x = dates, y = value, colour = country)) +
geom_line() + scale_colour_grey() + theme_minimal(base_size = 12) +
theme(legend.position = "none") + theme(panel.grid.minor = element_blank()) +
theme(axis.text.x = element_text(angle = 35, hjust = 1, color = "black", size = 9)) +
labs(x = NULL, y = "Cumulative incidence", title = "Confirmed Deaths", subtitle = NULL,
colour = NULL) + theme(plot.margin = unit(c(0.5,0.5,0.5,0.5), "cm")) +
scale_x_date(date_breaks = "2 weeks", date_minor_breaks = "2 weeks",
date_labels = "%Y-%m-%d", limits = c(as.Date("2020-02-26"),
as.Date("2020-05-20"))) +
scale_y_log10(breaks = 10**(1:10), labels = comma(10**(1:10)),
sec.axis = sec_axis(~ ., labels = NULL, name = "Log-scale")) +
theme(axis.title.y.right = element_text(angle = 90, size = 11),
axis.title.y.left = element_text(size = 11),
plot.caption = element_text(size = 9),
plot.title = element_text(hjust = 0.5)) +
annotate("text", y = df_deaths$value[which(df_deaths$date == "2020-05-15"
& df_deaths$country == "Argentina")] * 0.8,
x = as.Date("2020-05-17"), label = "Argentina") +
annotate("text", y = df_deaths$value[which(df_deaths$date == "2020-05-15"
& df_deaths$country == "Bolivia")] * 0.8,
x = as.Date("2020-05-17"), label = "Bolivia") +
annotate("text", y = df_deaths$value[which(df_deaths$date == "2020-05-15"
& df_deaths$country == "Brazil")] * 1.2,
x = as.Date("2020-05-17"), label = "Brazil") +
annotate("text", y = df_deaths$value[which(df_deaths$date == "2020-05-15"
& df_deaths$country == "Chile")] * 1.2,
x = as.Date("2020-05-17"), label = "Chile") +
annotate("text", y = df_deaths$value[which(df_deaths$date == "2020-05-15"
& df_deaths$country == "Colombia")] * 1.4,
x = as.Date("2020-05-17"), label = "Colombia") +
annotate("text", y = df_deaths$value[which(df_deaths$date == "2020-05-15"
& df_deaths$country == "Paraguay")] * 1.2,
x = as.Date("2020-05-17"), label = "Paraguay") +
annotate("text", y = df_deaths$value[which(df_deaths$date == "2020-05-15"
& df_deaths$country == "Peru")] * 1.2,
x = as.Date("2020-05-17"), label = "Peru") +
annotate("text", y = df_deaths$value[which(df_deaths$date == "2020-05-15"
& df_deaths$country == "Uruguay")] * 1.2,
x = as.Date("2020-05-17"), label = "Uruguay")
## Dataframe Recovered
df_recovered <- data.frame(dates = c(rep(as.Date(recovered_covid19$dates), 8)),
value = c(cumulate(recovered_covid19$ARG),
cumulate(recovered_covid19$BOL),
cumulate(recovered_covid19$BRA),
cumulate(recovered_covid19$CHL),
cumulate(recovered_covid19$COL),
cumulate(recovered_covid19$PRY),
cumulate(recovered_covid19$PER),
cumulate(recovered_covid19$URY)),
country = c(rep(arg_name, length(recovered_covid19$dates)),
rep(bol_name, length(recovered_covid19$dates)),
rep(bra_name, length(recovered_covid19$dates)),
rep(chl_name, length(recovered_covid19$dates)),
rep(col_name, length(recovered_covid19$dates)),
rep(pry_name, length(recovered_covid19$dates)),
rep(per_name, length(recovered_covid19$dates)),
rep(ury_name, length(recovered_covid19$dates))))
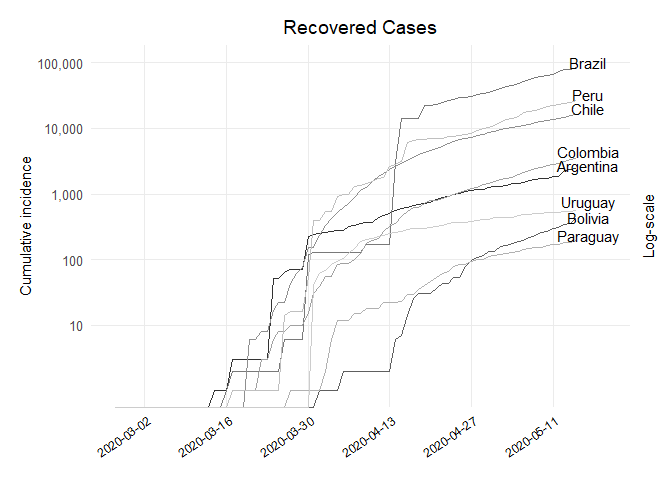
## Plot Recovered
ggplot(df_recovered, aes(x = dates, y = value, colour = country)) +
geom_line() + scale_colour_grey() + theme_minimal(base_size = 12) +
theme(legend.position = "none") + theme(panel.grid.minor = element_blank()) +
theme(axis.text.x = element_text(angle = 35, hjust = 1, color = "black", size = 9)) +
labs(x = NULL, y = "Cumulative incidence", title = "Recovered Cases", subtitle = NULL,
colour = NULL) + theme(plot.margin = unit(c(0.5,0.5,0.5,0.5), "cm")) +
scale_x_date(date_breaks = "2 weeks", date_minor_breaks = "2 weeks",
date_labels = "%Y-%m-%d", limits = c(as.Date("2020-02-26"),
as.Date("2020-05-20"))) +
scale_y_log10(breaks = 10**(1:10), labels = comma(10**(1:10)),
sec.axis = sec_axis(~ ., labels = NULL, name = "Log-scale")) +
theme(axis.title.y.right = element_text(angle = 90, size = 11),
axis.title.y.left = element_text(size = 11),
plot.caption = element_text(size = 9),
plot.title = element_text(hjust = 0.5)) +
annotate("text", y = df_recovered$value[which(df_recovered$date == "2020-05-15"
& df_recovered$country == "Argentina")]
* 1.1, x = as.Date("2020-05-17"), label = "Argentina") +
annotate("text", y = df_recovered$value[which(df_recovered$date == "2020-05-15"
& df_recovered$country == "Bolivia")] * 1,
x = as.Date("2020-05-17"), label = "Bolivia") +
annotate("text", y = df_recovered$value[which(df_recovered$date == "2020-05-15"
& df_recovered$country == "Brazil")] * 1.2,
x = as.Date("2020-05-17"), label = "Brazil") +
annotate("text", y = df_recovered$value[which(df_recovered$date == "2020-05-15"
& df_recovered$country == "Chile")] * 1.2,
x = as.Date("2020-05-17"), label = "Chile") +
annotate("text", y = df_recovered$value[which(df_recovered$date == "2020-05-15"
& df_recovered$country == "Colombia")] * 1.3,
x = as.Date("2020-05-17"), label = "Colombia") +
annotate("text", y = df_recovered$value[which(df_recovered$date == "2020-05-15"
& df_recovered$country == "Paraguay")] * 1.2,
x = as.Date("2020-05-17"), label = "Paraguay") +
annotate("text", y = df_recovered$value[which(df_recovered$date == "2020-05-15"
& df_recovered$country == "Peru")] * 1.2,
x = as.Date("2020-05-17"), label = "Peru") +
annotate("text", y = df_recovered$value[which(df_recovered$date == "2020-05-15"
& df_recovered$country == "Uruguay")] * 1.4,
x = as.Date("2020-05-17"), label = "Uruguay")
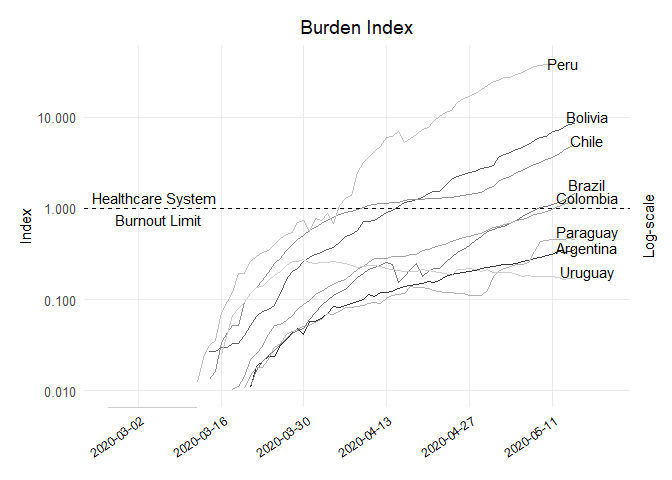
## Dataframe Burden Index (since 26 Feb to 15 may)
df_burden <- data.frame(dates = c(rep(as.Date(covid19$dates), 8)),
value = c(
(((cumulate(covid19$ARG) - cumulate(recovered_covid19$ARG))
* 0.15)/(icu_arg * 0.25)),
(((cumulate(covid19$BOL) - cumulate(recovered_covid19$BOL))
* 0.15)/(icu_bol * 0.25)),
(((cumulate(covid19$BRA) - cumulate(recovered_covid19$BRA))
* 0.15)/(icu_bra * 0.25)),
(((cumulate(covid19$CHL) - cumulate(recovered_covid19$CHL))
* 0.15)/(icu_chl * 0.25)),
(((cumulate(covid19$COL) - cumulate(recovered_covid19$COL))
* 0.15)/(icu_col * 0.25)),
(((cumulate(covid19$PRY) - cumulate(recovered_covid19$PRY))
* 0.15)/(icu_pry * 0.25)),
(((cumulate(covid19$PER) - cumulate(recovered_covid19$PER))
* 0.15)/(icu_per * 0.25)),
(((cumulate(covid19$URY) - cumulate(recovered_covid19$URY))
* 0.15)/(icu_ury * 0.25))
),
country = c(rep(arg_name, length(covid19$dates)),
rep(bol_name, length(covid19$dates)),
rep(bra_name, length(covid19$dates)),
rep(chl_name, length(covid19$dates)),
rep(col_name, length(covid19$dates)),
rep(pry_name, length(covid19$dates)),
rep(per_name, length(covid19$dates)),
rep(ury_name, length(covid19$dates))))
## Plot Burden Index
ggplot(df_burden, aes(x = dates, y = value, colour = country)) +
geom_line() + scale_colour_grey() +
geom_hline(yintercept = 1, col = "black", lty = 2) +
theme_minimal(base_size = 12) + theme(legend.position = "none") +
theme(panel.grid.minor = element_blank()) +
theme(axis.text.x = element_text(angle = 35, hjust = 1, color = "black", size = 9)) +
labs(x = NULL, y = "Index", title = "Burden Index", subtitle = NULL, colour = NULL) +
theme(plot.margin = unit(c(0.5,0.5,0.5,0.5), "cm")) +
scale_x_date(date_breaks = "2 weeks", date_minor_breaks = "2 weeks",
date_labels = "%Y-%m-%d", limits = c(as.Date("2020-02-26"),
as.Date("2020-05-20"))) +
scale_y_log10(limits = c(1e-2,4e1), labels = scales::comma,
sec.axis = sec_axis(~ ., labels = NULL, name = "Log-scale")) +
theme(axis.title.y.right = element_text(angle = 90, size = 11),
axis.title.y.left = element_text(size = 11),
plot.caption = element_text(size = 9),
plot.title = element_text(hjust = 0.5)) +
annotate("text", y = df_burden$value[which(df_burden$date == "2020-05-15"
& df_burden$country == "Argentina")] * 1.05,
x = as.Date("2020-05-17"), label = "Argentina") +
annotate("text", y = df_burden$value[which(df_burden$date == "2020-05-15"
& df_burden$country == "Bolivia")] * 1.2,
x = as.Date("2020-05-17"), label = "Bolivia") +
annotate("text", y = df_burden$value[which(df_burden$date == "2020-05-15"
& df_burden$country == "Brazil")] * 1.25,
x = as.Date("2020-05-17"), label = "Brazil") +
annotate("text", y = df_burden$value[which(df_burden$date == "2020-05-15"
& df_burden$country == "Chile")] * 1.1,
x = as.Date("2020-05-17"), label = "Chile") +
annotate("text", y = df_burden$value[which(df_burden$date == "2020-05-15"
& df_burden$country == "Colombia")] * 1.1,
x = as.Date("2020-05-17"), label = "Colombia") +
annotate("text", y = df_burden$value[which(df_burden$date == "2020-05-15"
& df_burden$country == "Paraguay")] * 1.2,
x = as.Date("2020-05-17"), label = "Paraguay") +
annotate("text", y = df_burden$value[which(df_burden$date == "2020-05-11"
& df_burden$country == "Peru")] * 1,
x = as.Date("2020-05-13"), label = "Peru") +
annotate("text", y = df_burden$value[which(df_burden$date == "2020-05-15"
& df_burden$country == "Uruguay")] * 1.2,
x = as.Date("2020-05-17"), label = "Uruguay") +
annotate("text", y = 1, x = as.Date("2020-03-05"),
label = "Healthcare System \n Burnout Limit")