COVID-19
COVID-19 Pandemic in South America
View the Project on GitHub bgonzalezbustamante/COVID-19-South-America
Estimated R and Explored Serial Interval Distribution in Paraguay
This online tracker was deprecated in mid-May 2020 to focus on a paper which evaluates early projections and governmental responses to COVID-19 epidemic in South America. This paper was published in World Development journal (DOI: 10.1016/j.worlddev.2020.105180). See its demonstration for R.
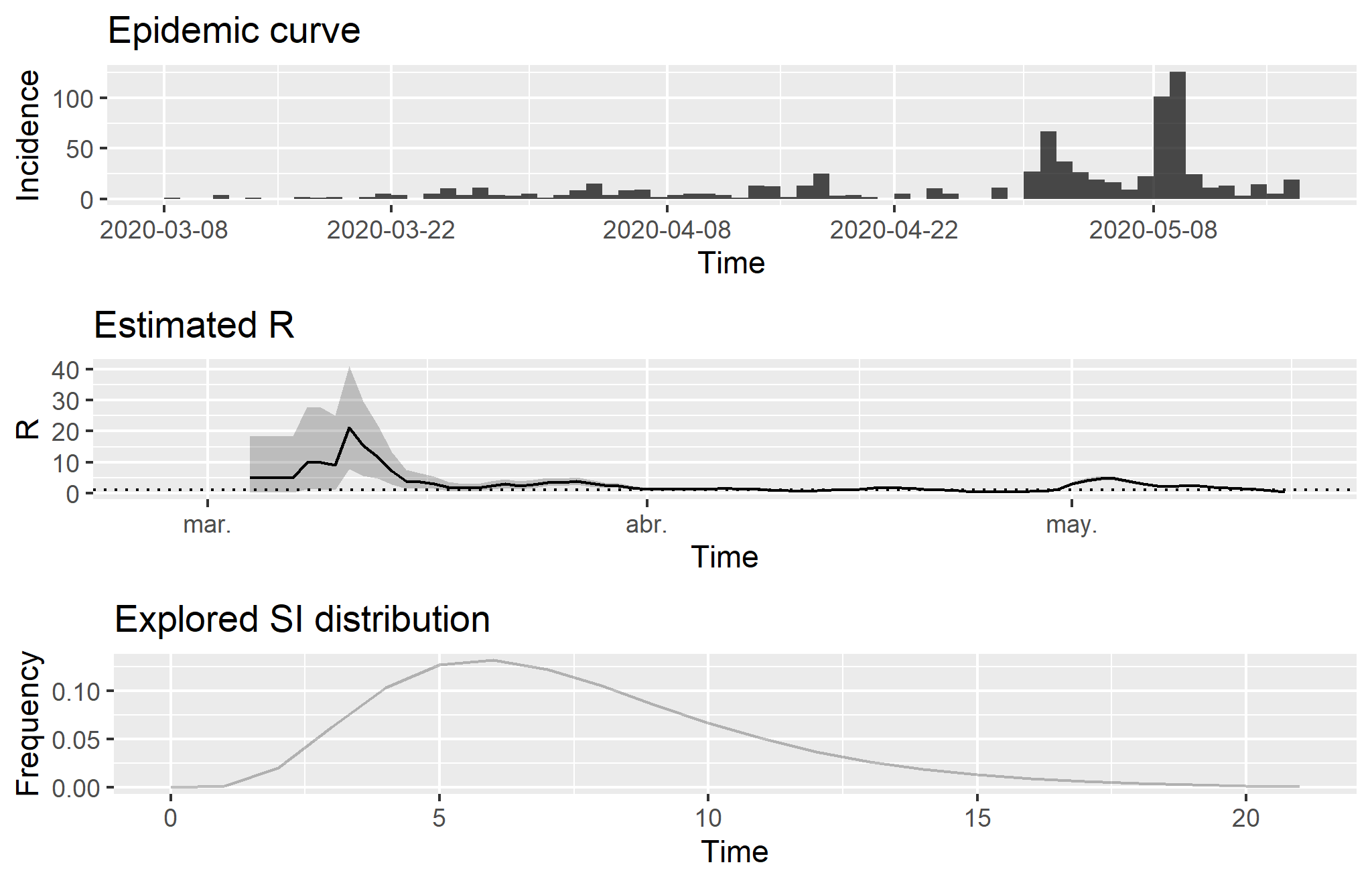 Source: Compiled using data from the Johns Hopkins University CSSE COVID-19 Dataset.
Source: Compiled using data from the Johns Hopkins University CSSE COVID-19 Dataset.
Note: The estimations have been calculated with a parametric serial interval based on a Gamma distribution with mean μ = 7.5 and standard deviation σ = 3.4 following to Li et al. (2020). The epicurve shows new infections over the period. On the other hand, estimated R illustrates the number of infections that a single case generates: when the value is lower than one, the outbreak is under control (see figure in detail). R is specifically the effective reproduction number (Re), which is calculated on a daily incidence basis. Therefore, it allows us to measure the public health systems response (see Cori et al., 2013; see also Thomson et al., 2019). Finally, the explored serial interval (SI) distribution shows the time interval between the symptoms in each case and the onset of them in secondary cases because of transmission. It is relevant to note that these estimations are too early in this epidemic to obtain a reliable posterior coefficient of variation of the R indicator.
Epidemic Trajectory and Future Incidence Simulation
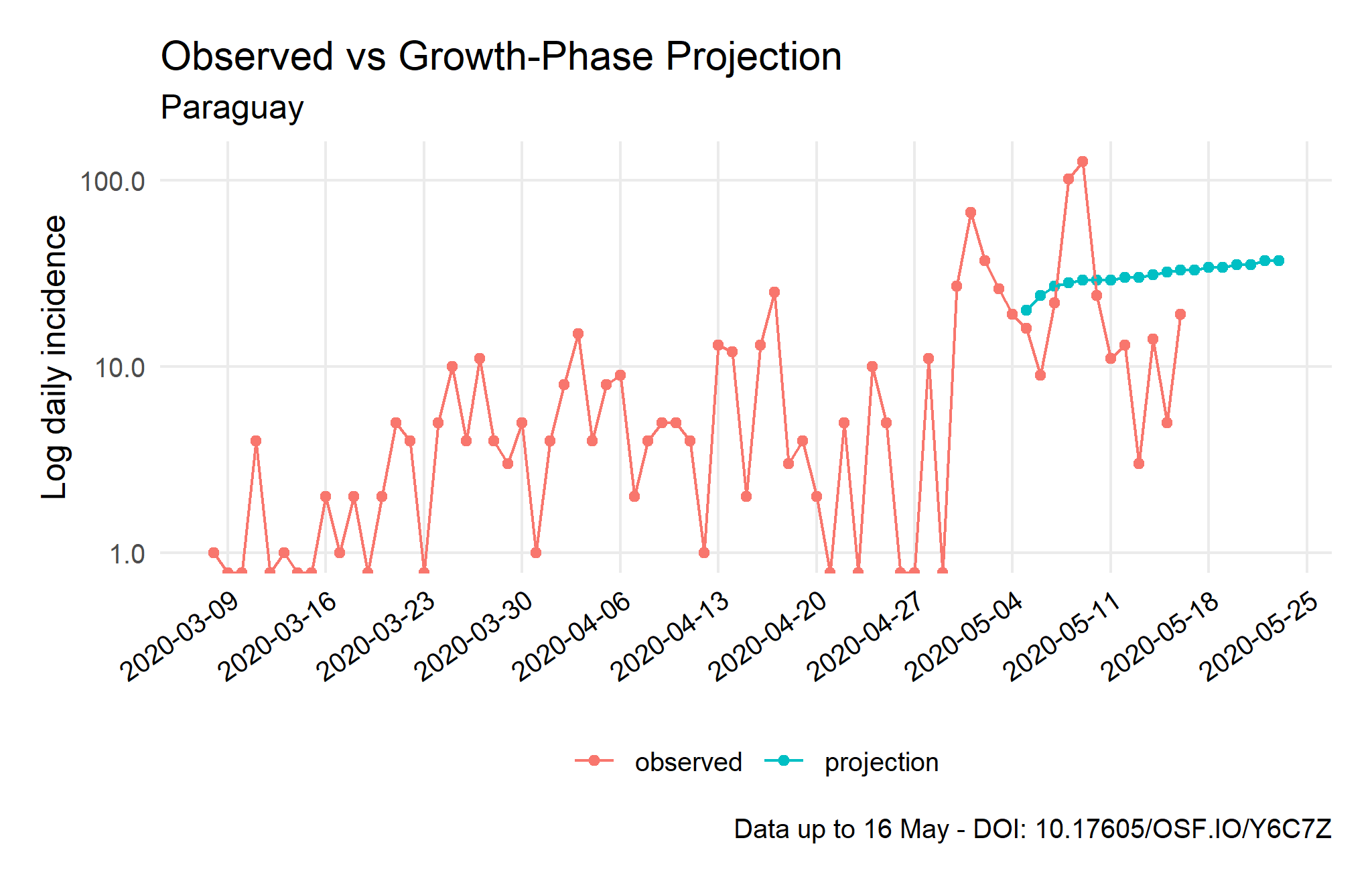 Source: Compiled using data from the Johns Hopkins University CSSE COVID-19 Dataset.
Source: Compiled using data from the Johns Hopkins University CSSE COVID-19 Dataset.
Note: The average number of secondary cases is calculated using a maximum-likelihood estimate of R based on the model of Cori et al. (2013) and the previous serial interval values. After that, 1,000 futures epicurves are simulated from the five days before the current peak, which is not necessarily the real growth-phase peak, to the next two weeks to compare observed and predicted values.
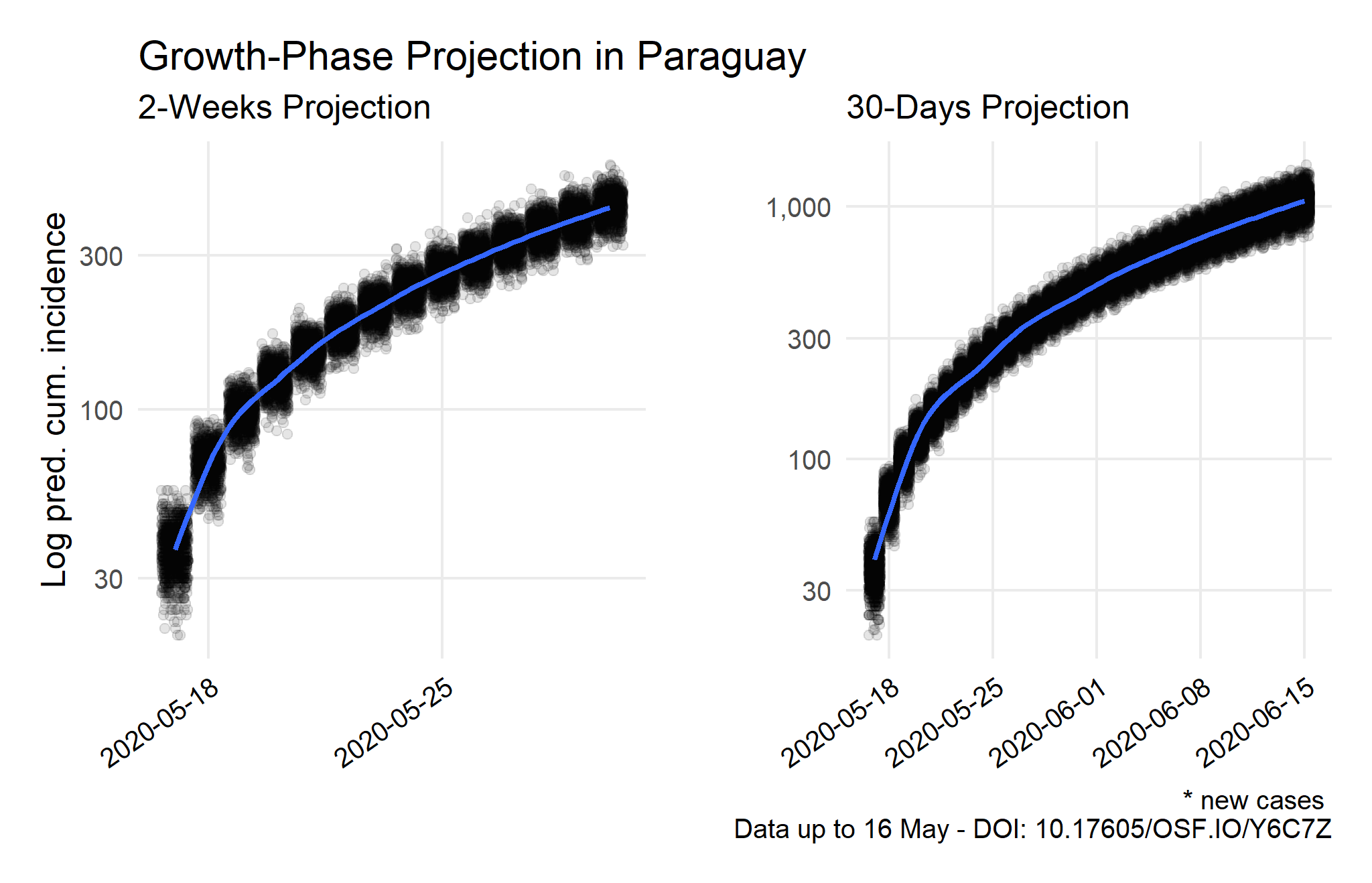 Source: Compiled using data from the Johns Hopkins University CSSE COVID-19 Dataset.
Source: Compiled using data from the Johns Hopkins University CSSE COVID-19 Dataset.
Note: The predicted cumulative incidence of new cases is estimated from the current peak to the following 30-days with 1,000 simulated epicurves per day based on the previous maximum-likelihood estimation and serial interval values. When the epidemic reaches a real peak, a log-incidence model should be estimated to measure the decay-phase adequately and to obtain the R daily growth rate in an advanced stage of the epidemic. This is a stronger assumption than a maximum-likelihood estimation, which is more useful to measure earlier stages. It is relevant to note this will increase the projections significantly in the growth phase. For further details, see the technical note.
Were You Looking for Previous Figures?
See the Visualisation COVID-19 component of the OSF-Project (DOI: 10.17605/OSF.IO/Y6C7Z)